This means the real secret for dairymen trying to develop naturally more immune, disease-resistant cows begins with genetic selection to enhance the profile of the 2,000 to 3,000 genes believed to control and direct the immune responses in animals.
Dr. Bonnie Mallard and the University of Guelph’s award-winning patented High Immune Response (HIR) technology is designed to identify cattle with optimized immune responsiveness and thereby greater resistance to a broad range of diseases.
The HIR technology is comprised of novel management and selection methods which reduce the negative impacts of disease and improve overall herd health and reproductive fitness.
As the technology matures and broadens in application with further advancements via genomics, it should have a profound and lasting impact on the health of cattle and profit on dairies.
Adaptive immune responses can be categorized into two types: antibody-mediated immune response (AMIR) and cell-mediated immune response (CMIR). AMIR reacts to infections outside of the cells within the body that are more commonly associated with bacterial-type infections such as mastitis, metritis and digital dermatitis.
CMIR is enacted to fight infections inside of the cells of the body that are typically caused by viral type infections or mycobacterial infections such as Johne’s disease and tuberculosis.
HIR combines these two types of adaptive immune responses for a more complete, broad-based defense against all types of bacterial and viral diseases.
This balanced approach between the two types of adaptive immune responses is critical because single-trait selection for one type (AMIR or CMIR) or for one specific disease can cause a negative impact with the resistance to other diseases.
High-immune-responder cows had about half the incidence of all diseases compared to low responders. Breeding to a high-immune-responder sire is expected to reduce all disease occurrences by 4 to 8 percent with each generation.
In addition to the benefit of disease reduction, selection for higher immune response will lead to offspring that respond better to commercial vaccines, thereby making a herd vaccination program more efficient and effective. It is estimated that up to 20 percent of animals have little or no response to commercial vaccination, in contrast to only 4 percent of high-immune responders.
Another major benefit is that research has shown how high-responder cows produce colostrum containing more specific antibodies (calving to five days postpartum) and elevated levels in the milk in late lactation.
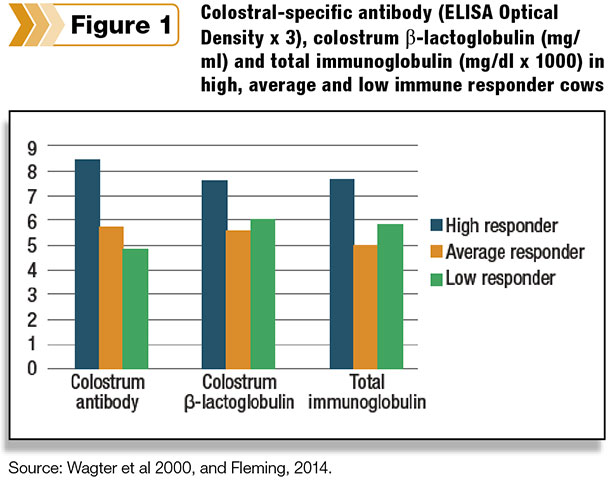
A recent study, with results shown in Figure 1, confirmed these findings by showing high-AMIR cows have greater concentrations of total Immunoglobulin and beta-lactoglobulin in colostrum compared with average and low responders. Immunoglobulin is the principal component of colostrum for passive immunity necessary for passing on early immunity benefits to newborn calves.
 The key figure in assessing the viability of a trait for making genetic progress is heritability. Heritability is defined as the portion of the variance observed in a population that is due to genetics, with the remainder of the variance influenced by management and environment.
The key figure in assessing the viability of a trait for making genetic progress is heritability. Heritability is defined as the portion of the variance observed in a population that is due to genetics, with the remainder of the variance influenced by management and environment.
Most health-related traits have low heritability estimates of less than 10 percent and some as low as 2 to 3 percent, which means the differences in cattle can be 97 to 98 percent explained by non-genetic sources.
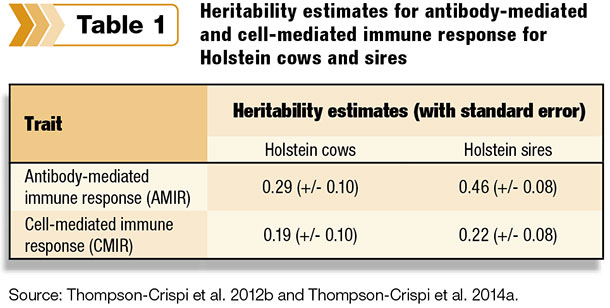
Two separate research projects, one on a population of females and one on a population of sires, showed immune response traits have a relatively high heritability (19 to 46 percent), which is comparable to the heritability for production traits (Table 1).
 It is feasible to make genetic gains in immune response relatively quickly in a way comparable to the genetic gains made by the dairy industry for production and highly heritable type traits.
It is feasible to make genetic gains in immune response relatively quickly in a way comparable to the genetic gains made by the dairy industry for production and highly heritable type traits.
Two separate genome-wide association studies for immune response were conducted, one based on a selected population of tested females and another on a larger group of non-related tested sires. The animals were genomics-tested using the Illumina 50K SNP chip and immune-tested using the HIR technology.
In the female study, 186 genetic markers were shown to differ between these cows based on antibody responses, and 21 genetic markers were associated with cell-mediated responses.
In the male study, 266 unique genetic markers were shown to differ between these sires based on antibody responses, and 46 unique genetic markers were significant in explaining differences between sires based on cell-mediated responses.
Genetic pathways included those within the bovine major histocompatibility complex, a complex gene region known to be important in the initiation and regulation of adaptive immune responses.
Mallard’s group at the University of Guelph is currently working to establish a large reference population of Holstein sires (about 2,000) and cows (about 3,000) with immune response phenotypes and 50K SNP genotypes. If successful, it may be possible in the near future to identify high- or low-immune responders from a simple hair sample.
Genomic-testing young heifers for immune response could become a key segment of a herd’s health management program. Similar to other heifer genomic-testing strategies, it could allow dairies to accentuate the high-immune-response heifers and to cull or reduce the contribution from low-immune-response heifers.
One can imagine herd strategies where high-immune-response cows become the basis for all colostrum collected and stored on farm.
Modifying management practices in the future based on knowledge of the animals’ immune response level may become a real possibility.
In the genomic era, the harmonization and complementary nature of genetics and management will have increasing importance for optimizing herd performance. PD
Dr. Bonnie Mallard is with the Ontario Veterinary College, Department Pathobiology, University of Guelph.
References omitted due to space but are available upon request. Click here to email an editor.

-
Jay Shannon
- Global Dairy Solutions Manager
- Semex
- Email Jay Shannon






